
Join NESM for our annual Fall Symposium & Business Meeting on Thursday, December 3rd at the Whitehead Institute in Cambridge, MA! The meeting will consist of four technical talks, a forty minute round of lightning talks, a coffee break, a buffet dinner, and a business meeting in which elections will be held for several positions on the Board of Directors. Election ballots will be distributed with the Fall E-Newsletter.
We are still accepting abstracts for one of five lightning talks!
Deadline: Wednesday, November 25 at 5pm
To submit an abstract go to:
http://nesmicroscopy.org/lightning-talk-submission/
Meeting Schedule
1:00 PM – Registration
1:30 PM – Welcoming - Jennifer Ross, PhD
1:40 PM – Vinothan Manoharan, Ph.D
2:20 PM – Gary Yellen, Ph.D
3:00 PM – Coffee Break
3:30 PM – Lightning Talks
4:10 PM – Discussion Break
4:40 PM – Nikta Fakrhi, Ph.D
5:20 PM – Tomas Kirchhausen, Ph.D
6:00 PM – Buffet Dinner
7:00 PM – Business Meeting
8:00 PM – Closing Remarks - Jennifer Ross, PhD
Additional Speaker Information
Vinothan Manoharan, Ph.D, Harvard University
"Looking at viruses using fiber optics and holograms"
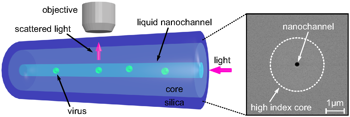
Abstract:
Simple viruses consist of RNA surrounded by a protective protein shell. Many of them are self-assembled: you can mix the RNA and the proteins in a test tube, and they will spontaneously form infectious viruses (don't worry -- none of the viruses I will discuss will infect you, unless you're a plant or bacterium). We want to understand how the virus manages to pull this off. But studying the dynamics of virus self-assembly is difficult, because the viruses are tiny (a few tens of nanometers in diameter), and they assemble quickly (taking somewhere between a microsecond and a second). I will discuss optical microscopy techniques that we have developed to be able to see these tiny things and resolve their dynamics on microsecond time scales.
Bio:
Vinothan N. Manoharan is the Wagner Family Professor of Chemical Engineering and Professor of Physics at Harvard University. His research focuses on understanding how systems containing many particles suspended in a liquid -- such as nanoparticles, proteins, or cells -- organize themselves into ordered structures like crystals, viruses, and even living tissues. His lab uses optical microscopy and holography to watch these systems self-assemble in real time. The goal is to discover new, general physical principles that underlie complex systems and to apply these principles to practical problems in materials science, nanotechnology, and medicine. Manoharan received his Ph.D. from the University of California, Santa Barbara in 2004 and worked as a postdoctoral researcher at the University of Pennsylvania before arriving at Harvard in 2005.
Gary Yellen, Ph.D, Harvard Medical School
"Quantitative imaging of genetically-encoded metabolic sensors in brain using FLIM"
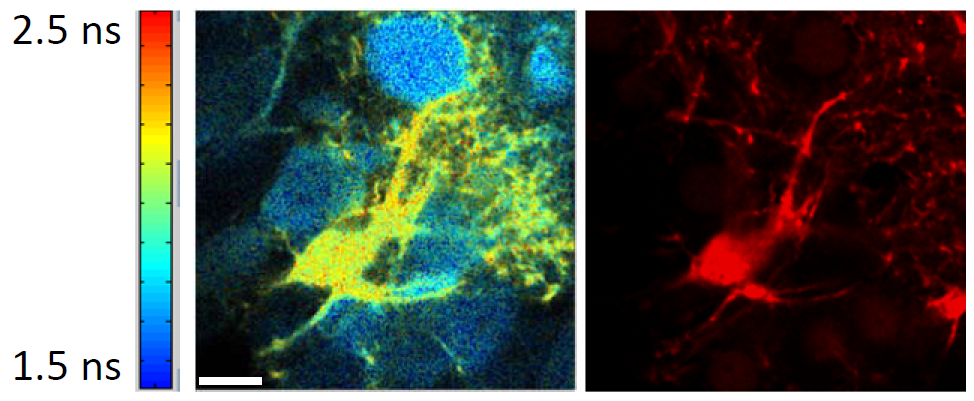
Abstract:
Although it is often considered as a mere “housekeeping” function of cells that presumably takes care of itself, cellular metabolism responds dynamically to energy demand and to changes in fuel supply, with important consequences for cellular function. It is now appreciated that metabolic reprogramming is key for turning normal cells into cancer cells, and this provides clues for effective cancer treatment. In the nervous system, there are equally dramatic effects of metabolism: drug-resistant epilepsy can be effectively treated using a dietary therapy that changes the fuel mixture burned by brain cells. To understand the dynamics of metabolism in living cells, we have developed several genetically-encoded fluorescent biosensors for key metabolites – ATP and NADH – and are imaging these in neurons and astrocytes, both in acute brain slices and in vivo. For two-photon quantitative imaging of the analytes (independent of the actual biosensor concentration), we use either ratiometric or fluorescence lifetime (FLIM) imaging. For several sensors with a good lifetime change, high speed FLIM provides calibrated, dynamic information about neuronal responses, on the timescale of seconds.
Bio:
Dr. Gary Yellen is Professor of Neurobiology at Harvard Medical School. He obtained his Ph.D. from Yale, working with C.F. Stevens, and did his postdoctoral research at Brandeis with Christopher Miller. For many years, his lab focused on understanding the "moving parts" of ion channel proteins. More recently, the lab has been working to understand the relationship between neuronal metabolism and excitability, with the ultimate goal of improving epilepsy treatment. Dr. Yellen’s interest in this area was piqued by the impressive success of a dietary therapy for epilepsy, the low carbohydrate “ketogenic diet”, which is still employed to treat the many people whose seizures do not respond to any of the existing antiseizure medications. The lab uses electrophysiology, mouse models, metabolic biosensors, and live cell imaging to discover how neuronal excitability responds to metabolic changes, and to explore the apparent role of the metabolically sensitive KATP channels found in many central neurons.
Nikta Fakrhi, Ph.D, MIT
"High resolution mapping of dynamic steady state transitions in actin cortices using single-walled carbon nanotubes"
Abstract:
Most animal cells are enveloped by a thin layer of actin cortex which governs the cell mechanics. A functional cortex must be rigid to provide mechanical stability while being flexible to allow for rapid restructuring such as during cell division. To satisfy these requirements, the actin cortex is highly dynamic with fast actin turnover in addition to myosin motors which generate active internal stresses. The regulatory mechanisms responsible for transitioning from stable steady states into restructuring states are not well understood. Here we develop a technique to map the dynamics of reconstituted actin cortices in emulsion droplets using IR fluorescent single-walled carbon nanotubes (SWNTs). We find that the cortex transitions between different dynamic steady states with an increase in crosslinkers concentration from a homogeneous state to one with rotational broken symmetry to a state with translational broken symmetry at high crosslinking. We apply the new dynamic mapping technique to cortices of live starfish oocytes in various developmental stages.
Bio:
Nikta Fakhri is an Assistant Professor in the Department of Physics at MIT. She completed her undergraduate degree at Sharif University of Technology, Tehran, Iran and her PhD at Rice University. She was a Human Frontier Science Program postdoctoral fellow at Georg-August-Universität in Göttingen, Germany before joining MIT in 2015. Her research focuses on non-equilibrium physics in biology. Active processes in living matter create a novel class of non-equilibrium materials composed of many interacting parts that individually consume energy and collectively generate motion or mechanical stresses. Active biological systems exhibit a wealth of intriguing properties, including anomalous fluctuations, non-equilibrium phase transitions, pattern formation on mesoscopic scales and unusual mechanical and rheological properties. Fakhri lab develops the use of fluorescent single-walled carbon nanotubes to probe the non-equilibrium properties of biological systems spanning a large range of length scales, from molecules to the cytoskeleton of individual cells, to tissues and whole organisms.
Tomas Kirchhausen, Ph.D, Harvard Medical School & Boston Children's Hospital
"Cellular Dynamics imaged in real time and in 3D using a lattice light sheet microscope"
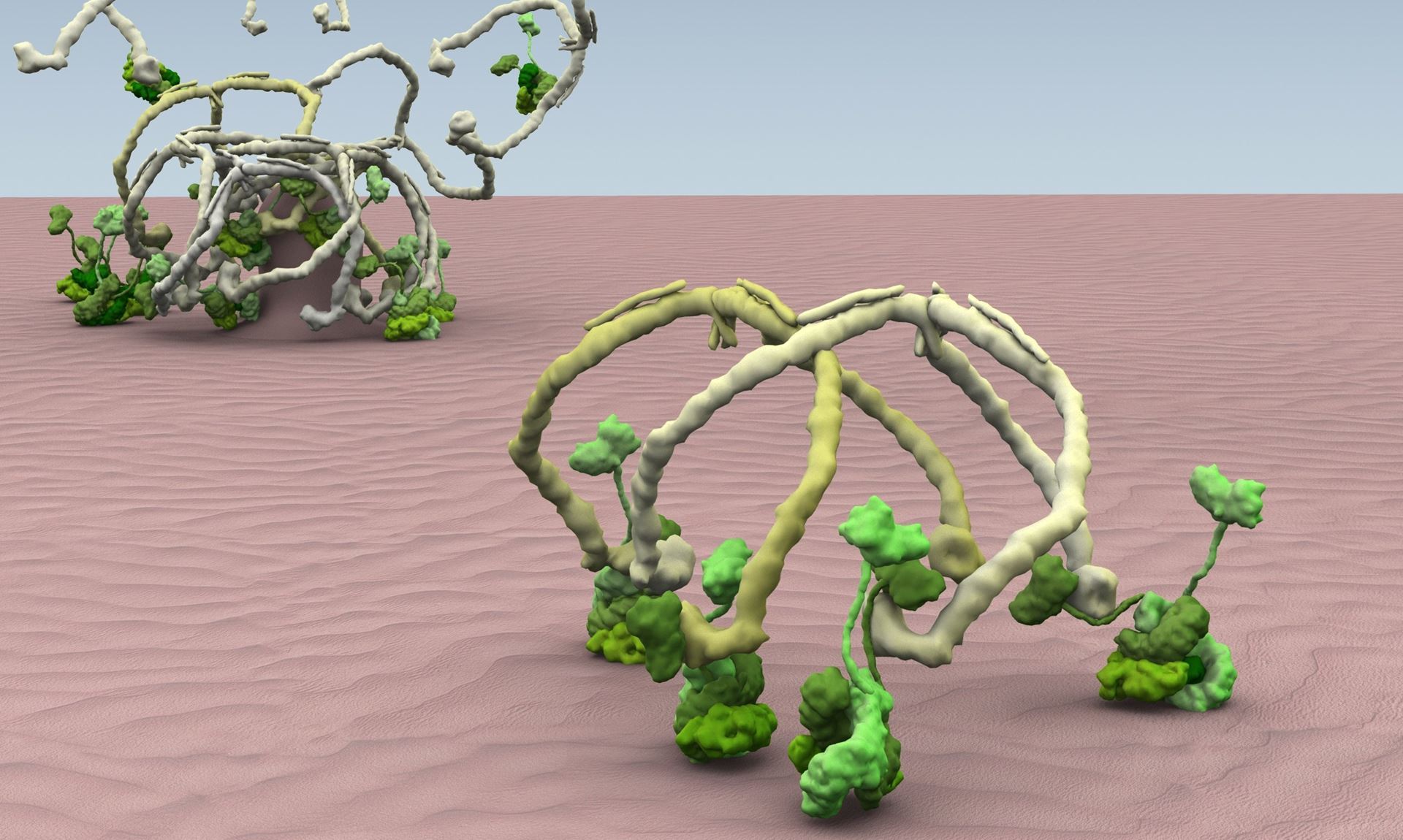
Abstract:
Our research focuses the processes that mediate and regulate the movement of vesicular carriers throughout cells and the biogenesis of organelles. In particular we study the molecular mechanisms that underlie the cell's sorting machineries responsiblndocytosis and for secretion, and how they are high jacked by toxins, viruses and bacterial pathogens to enter cells. We also study how organelles form and how during cell division, cells control their size and organelle architecture. The talk will illustrate our most recent efforts utilizing contemporary imaging tools based on live-cell fluorescence microscopy with single molecule sensitivity with high temporal resolution and spatial precision, to ‘see’ in three dimensions molecular events at the plasma membrane responsible for the formation of clathrin-coated pits and coated vesicles -- a conserved "nano-machine" that generates intracellular vesicular carriers in all animals and plants. The talk will also illustrate use of these imaging techniques to study the biogenesis of nuclear pores, how non-enveloped viruses penetrate cells and how cells control size during cell division.
Bio:
I received my Ph.D. in Biophysics from the Instituto Venezolano de Investigaciones Cientificas and my post-doctoral training at Harvard where I am now Professor of Cell Biology. Our research focuses on the processes that mediate and regulate the movement of membrane proteins throughout cells. In particular our studies have help define molecular mechanisms that underlie the cell's sorting machineries mediated by the clathrin pathway, the principal route responsible for receptor-mediated endocytosis and for secretion, a route critical for reuptake of membrane at synapses, and a mode of entry usurped by many viral and bacterial pathogens. These biological phenomena have importance for the understanding of such diseases as cancer, viral infection and pathogen invasion, Alzheimer's, as well as other neurological diseases. We also study how during cell division, cells control their size and organelle architecture. Our work has been characterized by use of emerging technologies -- from the early days of molecular cloning to contemporary live-cell imaging. We use the tools of x-ray crystallography, NMR, electron cryomicroscopy, and single-molecule biophysics to create a "molecular movie" of clathrin-mediated endocytosis, and in this way relate these molecular events to functional properties of the surfaces of living cells. We have helped defined the structure, interactions, and assembly-disassembly mechanisms of clathrin and many of its associated proteins, through studies extending over three decades, including the first X-ray crystal structure determination of clathrin and the AP-1 endosomal clathrin adaptor, and the first high resolution cryo-electron microscopy of a complete clathrin coat. We also use frontier optical-imaging modalities including super-resolution total internal reflection fluorescence microscopy and lattice light sheet fluorescence microscopy with single molecule sensitivity. Our fluorescent high resolution imaging microscopy techniques are geared to gather in 3D and real time, quantitative, “single-molecule” and "single-object" data to examine cellular membrane remodeling processes from in vitro reconstituted systems or from living cells. Focusing on clathrin-coated pits and coated vesicles, for example, we tracked them while they are assembling, recruiting cargo, membrane scission, budding and uncoating. The fluorescent probes include clathrin, AP-2, auxilin, dynamin and other molecules expressed in gene-edited cells as well as fluorescently tagged cargo. With these type of studies we integrate molecular snapshots obtained at high resolution with real time in vitro and live-cell processes, to generate ‘molecular movies' aimed towards obtaining new frameworks for analyzing some of the molecular contacts and switches that participate in the regulation, availability, and intracellular traffic of the many molecules involved in signal transduction, immune responsiveness, lipid homeostasis, cell-cell recognition and organelle biogenesis
Location
McGovern Auditorium
Whitehead Institute
9 Cambridge Center
Cambridge, MA 02142
Parking
The Whitehead Institute is in the heart of Kendall Square/MIT and is within easy access of the Red Line T and bus routes.
- Traffic can be tough in Kendall Square so, if you decide to drive, please check Google directions to allow sufficient time.
- The map at http://en.parkopedia.com/parking/kendall_sq_cambridge/ shows local parking options. Whitehead Institute is located at the intersection marked with the location pin and the metered street parking pin.
- Street parking is limited to two hours and Cambridge Police are diligent in enforcing that.