
You are invited to the New England Society for Microscopy's 33rd Annual Spring Workshops & Symposium at the Marine Biological Laboratory in Woods Hole, Massachusetts! The meeting will be held over two days - Thursday, April 28th, will feature two workshops which will allow a more in-depth exploration of state-of-the-art microscopy techniques. Registration and information about the Workshops can be found here. Friday, April 29th, will consist of a day-long Symposium featuring four technical talks, six 6-minute "lightning" talks, a poster session, an exhibiting vendor session, and a buffet lunch.
We are currently accepting abstract submissions for poster sessions or lightning talks. Please submit abstracts here by April 18th.
Schedule
9:00 AM – Registration & Continental Breakfast
10:00 AM – Welcoming Remarks
10:10 AM – "High-Speed Serial Electron Microscopy using a 61 Beam Scanning Electron Microscope" Richard Schalek, Harvard University
10:50 AM – "Lightning Talks"
11:30 AM – Poster & Vendor Session
12:30 PM – Buffet lunch
1:45 PM – "Hard Microscopy Experiments on Soft Materials: Insights and Opportunities with Surfaces, Cells, and Membranes"
Maria Santore, PhD, UMass Amherst
2:25 PM – Afternoon break
3:00 PM – "Quantifying the dynamics of establishment and maintenance of cell cycle arrest in individual cells using fluorescence time-lapse imaging" Jose Reyes, Harvard Medical School
3:40 PM – "Dynamic Reorganization of Nanoporous Gold Catalysts During Selective Oxidation Reactions" Branko Zugic, PhD,Harvard University
4:20 PM – Closing
Speaker Abstracts & Bios
"High-Speed Serial Electron Microscopy using a 61 Beam Scanning Electron Microscope"
Richard Schalek, Harvard University
Abstract:
The cellular organization of the mammalian brain with length scales spanning centimeters to nanometers is more complicated than that of any other known biological tissue. The precise synaptic relationships between the neuronal circuits that interconnect thousands of neurons is largely a mystery. By imaging volumes of brain using 3-dimensional electron microscopy, the details of the neuronal shape and connectivity can be reconstructed. The recent technological advance of the automated tape ultramicrotome (ATUMtome) has automated the collection of serial sections so that thousands of 30 nm thick sections can be collected with relative ease. Imaging these datasets requires that the dataset be of sufficient size and resolution to reconstruct the connectivity of the neurons composing a circuit –its wiring diagram or ‘connectome’ can be extracted. To move to the next length and volume scale of neuronal circuits requires several technological advances. The multibeam scanning electron microscope (mSEM) represents a transformative imaging technology that enables neuroscientists to tackle millimeter scale cortical circuit problems. In this work we describe a workflow from tissue harvest to imaging that will generate extremely --large petabyte datasets (300,000,000 images) of cortex.
Bio:
Richard Schalek is currently a Research Associate in Prof. Jeff Lichtman’s group in the Department of Molecular and Cellular Biology at Harvard University. Richard received a Bachelor of Science in Physics from Angelo State University, a Master degree in Physics from the University of North Texas and PhD in Materials Science from Michigan State University. After a post-doc in the Department of Mechanical Engineering at the University of Nebraska, Richard work as a Staff Scientist in the Composite Materials and Structure Center at Michigan State University. In 2002 Richard joined the Center for Nanoscale Systems at Harvard University as Leader of the Materials Group. Richard joined Prof. Lichtman’s group in 2007 to help develop technologies for high-speed large-volume imaging of cortical circuits. Richard has served as President, Treasurer and Secretary for New England Society for Microscopy and the Michigan Microscopy Society.
"Hard Microscopy Experiments on Soft Materials: Insights and Opportunities with Surfaces, Cells, and Membranes"
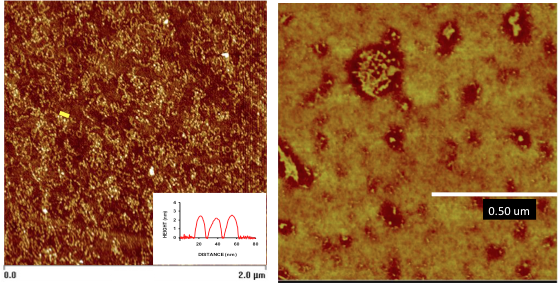
Maria Santore, PhD, UMass Amherst
Abstract:
Research in the Santore lab focuses on the creation of engineered surfaces with targeted interactions, which find ultimate use in a variety of applications, from microfluidic processing to controlled interactions with cells and tissues. In our quest for quantitative interpretation of experimental results, to facilitate mechanistic treatments of interfacial behaviors and to develop design rules towards commercial materials processing, we encounter recurring challenges in our microscopy studies. This talk provides examples of our successes, needs, and frustrations with different microscopy methods (AFM, single photon, TEM, particle tracking, and confocal imaging) and highlights how we combine microscopy with non-imaging methods in our research. In one example, we successfully quantify minute quantities of adsorbed fibrinogen on a surface (at these low levels, the adsorbed protein is sufficient to produce infection or other adhesive interactions), but we fail to achieve the needed quantitative resolution when the protein adsorbs in clusters on nanoparticle-bearing surfaces. In another example, engineered surfaces (patent pending) are capable of adhesively distinguishing epithelial cancer cells from white blood cells, but the driving force is unknown. Using microscopy we attempted to probe the interactions of targeting species with cell surfaces but the species were taken up so quickly that the dynamics prevented imaging. In a third example probing the tension-sensitivity of phase separation in phospholipid membranes, an inverted fluorescence microscope proves more effective than confocal microscopy because of the thermal membrane fluctuations compared with the slower timescale of obtaining the confocal image stack.
Bio:
Maria Santore is a Professor of Polymer Science and Engineering, with a secondary appointment Chemical Engineering at the University of Massachusetts at Amherst.
Santore was trained formally as a Chemical Engineer, obtaining her B.S. at Carnegie Mellon and Ph.D. at Princeton. Following her Ph.D., Santore’s postdoctoral studies of polymer glasses and blends in the Polymer Division at NIST in Gaithersburg MD were supported by an NRC Postdoctoral Fellowship.
She joined the Chemical Engineering Department at Lehigh University as the Dana Assistant Professor of Chemical Engineering and was promoted to Associate Professor in subsequent years. She was awarded the Alfred Nobel Robinson Award for outstanding research in 1996 and was named the Class of 1961 Chaired Associate Professor while at Lehigh. Following a sabbatical at the Institute for Medicine and Engineering at the University of Pennsylvania she joined the University of Massachusetts Department of Polymer Science and Engineering as a Full Professor.
Santore is currently a senior editor of Langmuir, the ACS (American Chemical Society) Journal of Surfaces and Colloids, and a section editor for the Elsevier Journal Current Opinion in Colloid and Interface Science. She is a Fellow in the American Physical Society, the American Association for the Advancement of Science, and the American Chemical Society. She was awarded the 2015 Hopper Lectureship in Engineering by the University of Pennsylvania. Her research focuses on dynamic aspects of bioadhesion, cell manipulation, and targeted delivery using polymers, biomacromolecules, colloids, and nanoparticles at surfaces.
"Quantifying the dynamics of establishment and maintenance of cell cycle arrest in individual cells using fluorescence time-lapse imaging"
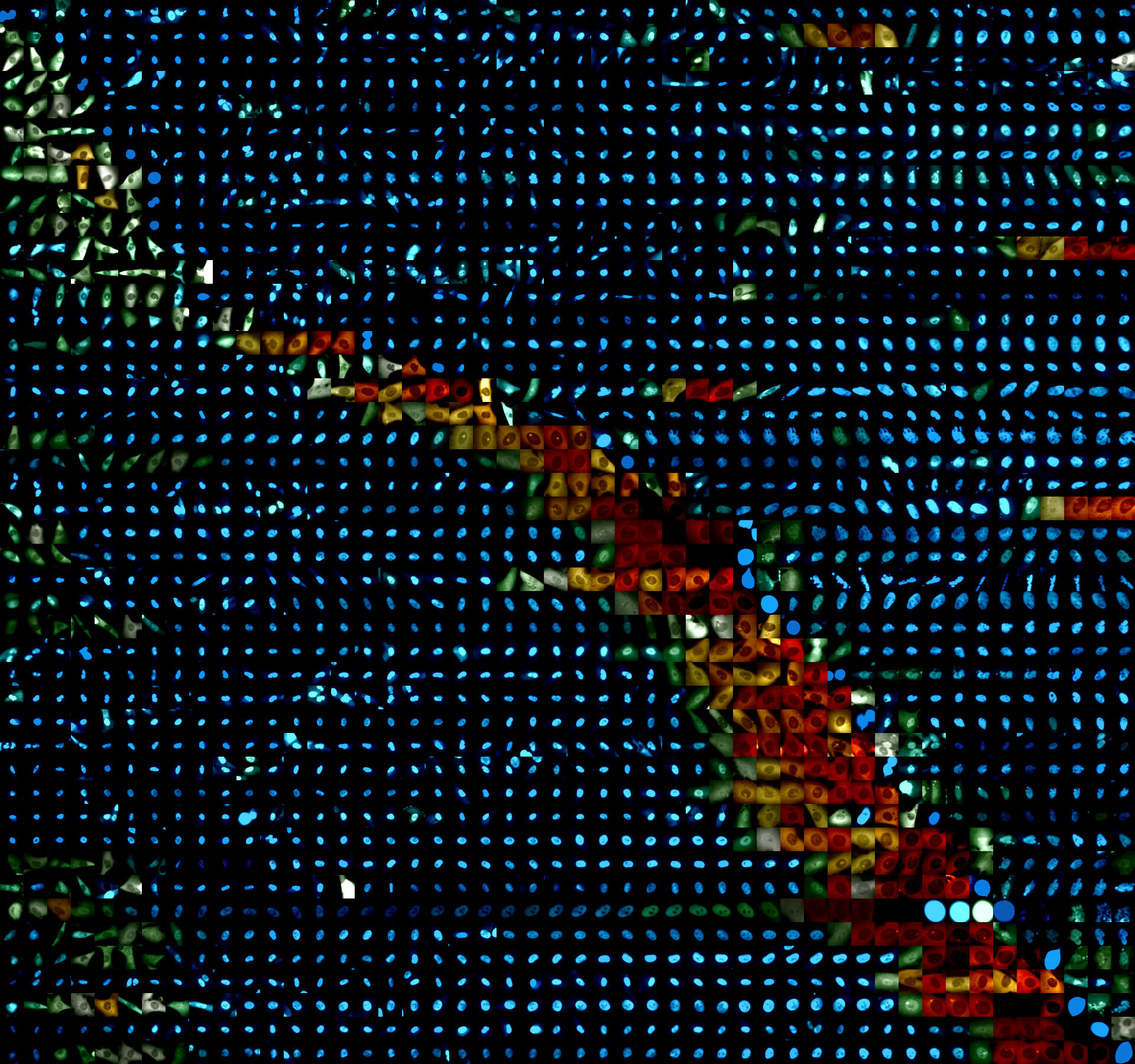
Jose Reyes, Harvard Medical School
Abstract:
Cellular senescence is a terminal cell fate characterized by the long-term maintenance of cell cycle arrest that results as an outcome of DNA damage, among other types of stress. In order to quantitatively understand the dynamics of establishment of this phenotypic state, we have used fluorescence time-lapse live cell imaging to track the division history of individual human cells for up to 7 days after DNA damage. We have identified a subpopulation of cells maintains a cell cycle arrest in the presence of DNA damage signaling for several days, but eventually escapes such state in sporadic events that are widely distributed in time. Using live cell reporters for p53 and p21, two important molecular players that link DNA damage sensing and cell cycle arrest, we have identified temporal signatures in the dynamical behavior of these proteins that correlate with cell cycle re-entry in these 'late dividing' cells. Our work suggests that fluctuations in DNA damage signaling can limit the long-term maintenance of cell cycle arrest in non-transformed cells. We are interested in understanding the mechanistic basis of such fluctuations and the way they can be perturbed to shape the stability and escape kinetics from cell cycle arrest.
Bio:
I did my undergraduate studies in the National Autonomous University of Mexico, where I conducted research aimed at understanding the extent and functional consequences of large-scale genomic rearrangements in healthy human individuals and cancer genomes. I then moved to the Systems Biology PhD program at Harvard University in 2011, where I joined Galit Lahav's lab. I’m very interested in understanding the way biological systems compute information from their environment and internal state, and translate this into the regulation of complex behavior. Today I will present recent insights stemming from our observations of the response of human cells over long timescales, and the way fluctuations in DNA damage signaling shape the long-term stability of cellular fate.
"Dynamic Reorganization of Nanoporous Gold Catalysts During Selective Oxidation Reactions"
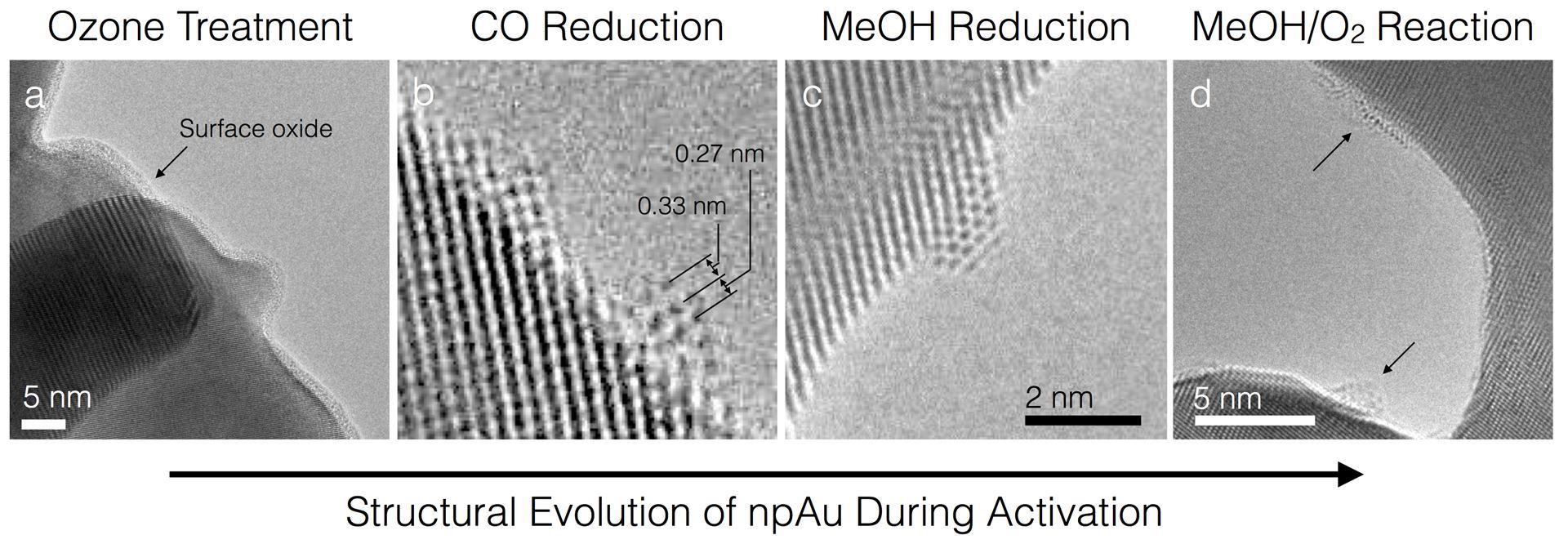
Branko Zugic, PhD, Harvard University
Abstract:
Bimetallic catalysts are critical to the development of tunable material properties in order to address energy inefficiencies in catalytic processes. The dynamic nature of these materials has often been overlooked in their design. Nanoporous gold (npAu) is an unsupported metallic Au alloy with dilute Ag content. It requires activation with ozone in order to selectively catalyze the O-assisted coupling of methanol and other alcohols. We have shown that treatment of npAu by ozone at 150 °C followed by exposure to a methanol-oxygen mixture results in highly active and stable catalysts. To understand the structural evolution and catalytic activity after ozone treatment, we have used a combination of reactor studies, theoretical calculations, atmospheric pressure X-ray photoelectron spectroscopy (AP-XPS), and electron microscopy (including environmental-TEM). The results show that dynamic structural changes take place at the npAu surface at different stages of the activation process. The ozone treatment causes near-complete oxidation of the npAu surface that results in amorphous layers of silver/gold oxides. This oxide layer gives rise to non-selective products (i.e. combustion). However, the ‘oxidic’ oxygen can be removed by CO titration, leading to metastable structures that are reactive selectively to methanol. These quasi-ordered structures appear to be responsible for the selective oxidation activity of npAu and are predicted by DFT studies. The results represent the potential of correlated in-situ/DFT studies and highlight their ability to improve our understanding of the dynamics of bimetallic catalysts.
Bio:
Branko studied chemical engineering at Worcester Polytechnic Institute (BS) and Tufts University (PhD). His PhD work focused on the development of alkali-promoted platinum nanocatalysts for the water-gas shift reaction and biomass-to-fuel processes. He joined the Friend lab at Harvard University in July 2013 and is currently investigating the use of nanoporous gold (npAu) for oxidation and coupling reactions. In addition to developing a reactor for evaluating the catalytic performance of npAu, Branko uses advanced in-situ/operando electron microscopy (TEM, STEM) and X-ray spectroscopy (XPS) tools to characterize and better understand the reactive and surface properties of such materials.
Location
Swope Conference Center
7 M B L Street
Woods Hole, MA
02543
Parking
Free parking facilities are located on-site. Upon arrival, please stop at the Swope Center to pick up a parking pass. Visitors will park at the Bar Neck lot.
Housing