Join NESM for our annual Fall Symposium & Business Meeting on Thursday, November 9th at the University of Massachusetts, Amherst, MA!
The meeting will consist of four or five technical talks, a poster session, a coffee break, a catered lunch, and a business meeting in which elections will be held for several positions on the Board of Directors. Election ballots will be distributed with the Fall E-Newsletter.
We will be celebrating the 50th Anniversary of our Society!
Nov 8th: Workshops at the new Light Microscopy Facility.
This facility is one of a few designated Nikon Centers of Excellence. Nikon specialists will be providing a unique opportunity for training and demonstrations of the diverse available microscope.
You can even bring your own samples to test!!!
Register for workshops HERE.
12-1pm: Check-in at room S576 Life Science Laboratories)
1-3pm: Workshop 1 (Super-resolution) and Workshop 2 (Scripting/JOBS)
3-5pm: Workshop 3 (Enhanced resolution) and Workshop 4 (High Content Imaging)
Nov 9th: Symposium
9am Registration at S330 Life Science Laboratories / 45 min tour of EM facility leaves LSL at 9:15
10am Welcome
10:10-10:50am Talk 1 Tomomi Tani
10:50-11:30am Talk 2 Bruce Goode
11:30-12:30pm Poster Session (Submit title/abstract here)
12:30-2pm NESM History/Lunch/Business Meeting
2-2:40pm Talk 3 Dan Needleman
2:40-3:20pm Talk 4 Jennifer Ross
3:30-4pm Closing Celebration
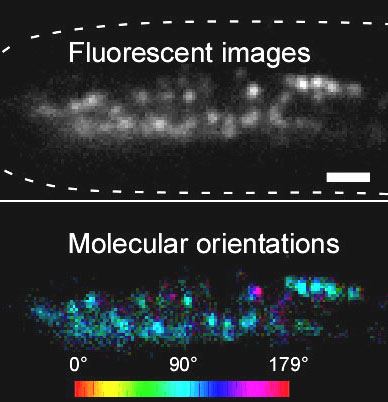
Talk 1: Tomomi Tani; Dissection of molecular assembly dynamics by tracking orientation and position of single molecules in live cells
Abstract 1: Regulation of molecular alignment drives the physiological functions of many protein assemblies in living cells yet remains difficult to explore accurately through space and time. We built an instantaneous fluorescence polarization microscope, which simultaneously images position and orientation of fluorophores in living cells with single-molecule sensitivity and a time resolution of 100ms. We developed image acquisition and analysis methods to track single particles that interact with higher-order assemblies of molecules. We tracked the fluctuations in position and orientation of molecules from the level of an ensemble of fluorophores down to single fluorophores. We tested our system in vitro using fluorescently labeled DNA and F-actin in which the ensemble orientation of polarized fluorescence is known. We then tracked the orientation of sparsely labeled F-actin networks at the leading edge of migrating human keratinocytes, revealing the anisotropic distribution of actin filaments relative to the local retrograde flow of the F-actin network. Additionally, we analyzed the position and orientation of septin-GFP molecules incorporated in septin bundles in growing hyphae of a filamentous fungus. Our data indicate that septin-GFP molecules undergo positional fluctuations within ~350 nm of the binding site and angular fluctuations within ~30° of the central orientation of the bundle. By reporting position and orientation of molecules while they form dynamic higher-order structures, our approach can provide new insights into how micron-scale ordered assemblies emerge from nano-scale molecules in living cells.

.png)
Talk 2: Bruce Goode; Actin polymerization throttles: a case solved by live-cell imaging and in vitro single molecule TIRF microscopy
Abstract 2: This talk will highlight our recent use of quantitative live-cell imaging and in vitro single molecule TIRF microscopy to define machinery and mechanisms controlling cell shape and movement. Each cell in our bodies is filled with a network of thousands of interconnected polymers (protein fibers) collectively called the cytoskeleton. The two primary cytoskeletal polymer systems are actin filaments and microtubules, and both can be rapidly assembled and reorganized to drive cell motility and cell morphogenesis. We started with a simple question: how do cells build linear actin structures (actin cables) of a particular length to fit into a cellular compartment of a particular size? We discovered that actin cables are assembled by a remarkable class of proteins, called Formins, which both ‘seed’ the formation of new actin filaments and accelerate filament growth to different degrees. This shows us that nanometer sized protein machines reside at one end of a micron-scale cytoskeletal fiber, governing its length (and shape) to match the geometry of a cell. Additionally, we found that in vivo binding partners of Formins can throttle the speed of actin filament growth up or down, and serve as components of feedback mechanisms controlling actin cable length, or harness actin polymerization to microtubules to coordinate the dynamics of the two polymer systems.
Bio 2: Bruce Goode is a Professor of Biology at Brandeis University. He earned his B.S. and his Ph.D. in Cell Biology from the University of California Santa Barbara, working with Stuart Feinstein. He conducted his postdoctoral research in the laboratory of David Drubin at the University of California Berkeley. In 2000, Dr. Goode started his lab at Brandeis, where his research program focuses on mechanisms of actin and microtubule cytoskeleton remodeling underlying cell motility, cell morphogenesis, and intracellular transport. Work in his lab is equally divided between yeast and mammalian systems, and projects are multi-disciplinary in nature, combining in vitro single molecule TIRF imaging, genetics, biochemistry, live cell imaging, and single particle EM. Dr. Goode has received scholar awards from the Pew Charitable Trust, March of Dimes, and American Cancer Society, and a Research Career Development Award from the NIH. He is on the F1000 advisory board and the editorial boards of The Journal of Cell Biology and Molecular Biology of the Cell. He was the Editor-in-Chief of Cytoskeleton from 2009-2016. His lab's research accomplishments include: (1) the discovery that formins polymerize actin; (2) elucidating novel mechanisms of formin regulation by its binding partners (e.g., APC, CLIP-170, Bud6, Bud14, Smy1, and Hof1); (3) describing roles for unconventional Arp2/3 complex regulators (e.g., Coronin, GMF, and Abp1); (4) uncovering mechanisms in the control of actin filament disassembly (e.g., by AIP1, Coronin, Twinfilin, and Srv2/CAP); and (5) defining the molecular basis of microtubule and actin coordination (e.g., by APC, CLIP-170, and Profilin).


Talk 3: Dan Needleman; Biophysics of Spindle Positioning and Elongation
Abstract 3: The spindle is positioned asymmetrically during the first mitotic division in C. elegans. We are investigating how different forces coordinated to move the spindle, and if these forces are generated from interactions with the cytoplasm, the cortex, or a combination of both. For this purpose, we constructed a laser ablation system capable of cutting complex patterns with high spatial and temporal precision, and we are applying it to quantitatively perturb spindle movements. We are also using fluorescent nanodiamonds to track cytoplasmic fluid flow as the spindle moves. We interpret our data using a combination of theory and simulations (in collaboration with Mike Shelley, NYU/Courant/Flatiron). Our results argue that pulling forces from the cortex drive key aspects of spindle motions, including the initial centering, subsequent asymmetric positioning, transverse oscillating behaviors, and elongation. We hope to provide a quantitative, integrated understanding of spindle positioning.

Talk 4: Jennifer Ross; How does the cell organize its insides?
Abstract 4: The cell is a complex autonomous machine taking in information, performing computations, and responding to the environment. To enable agile read/write capabilities, much of the molecular biochemistry that performs these computations must be transient and weak, allowing signals to be carried as a function of the concentration of numerous and coupled interactions. Traditionally, biochemical experiments can only measure strongly interacting systems that can last for long times in dilute concentrations. We have developed microscopy measurements to enable to visualization of weak, transient interactions and the resulting emergent behaviors of coupled systems. I will present excerpts from stories where many weak, transient interactions can have strong repercussions on the overall activity and can, in fact, overpower strongly interacting systems. These studies involve the microtubule cytoskeleton and the transport motor, kinesin-1. Our results reveal a fundamentally important aspect of cellular self-organization: weak, transient interacting species can tune their interaction strength directly by tuning the local concentration to act like a rheostat. The tunability of weak, transient interactions is a fundamental activity of biological systems, and our insights will ultimately enable us to learn how to engineer thesis systems to create biological or biomimetic devices.
Bio 4: Ross is the director of the new Massachusetts Center for Autonomous Materials (MassCAM) and an award-winning biophysicist studying the organization of the microtubule cytoskeleton and microtubule-based enzymes using high-resolution single molecule imaging techniques. She has a degree in Physics, and has studied the microtubule cytoskeleton for over a decade. As a Cottrell Scholar, Ross has pioneered innovative teaching techniques that are being adopted around the world. Specifically, ahe has taught at several international short courses on microscopy including Analytical and Quantitative Microscopy (AQLM) at the Marine Biology Laboratory and the Bangalore Microscopy Course at the National Centre for Biological Science in Bangalore, India. She has also served as the President of NESM in the past. She is also an advocate for women and under-represented groups and has a blog to help others make it in academics.

Location
UMass, Amherst
S330/340 Life Science Laboratories
240 Thatcher Way
Amherst, MA 01003
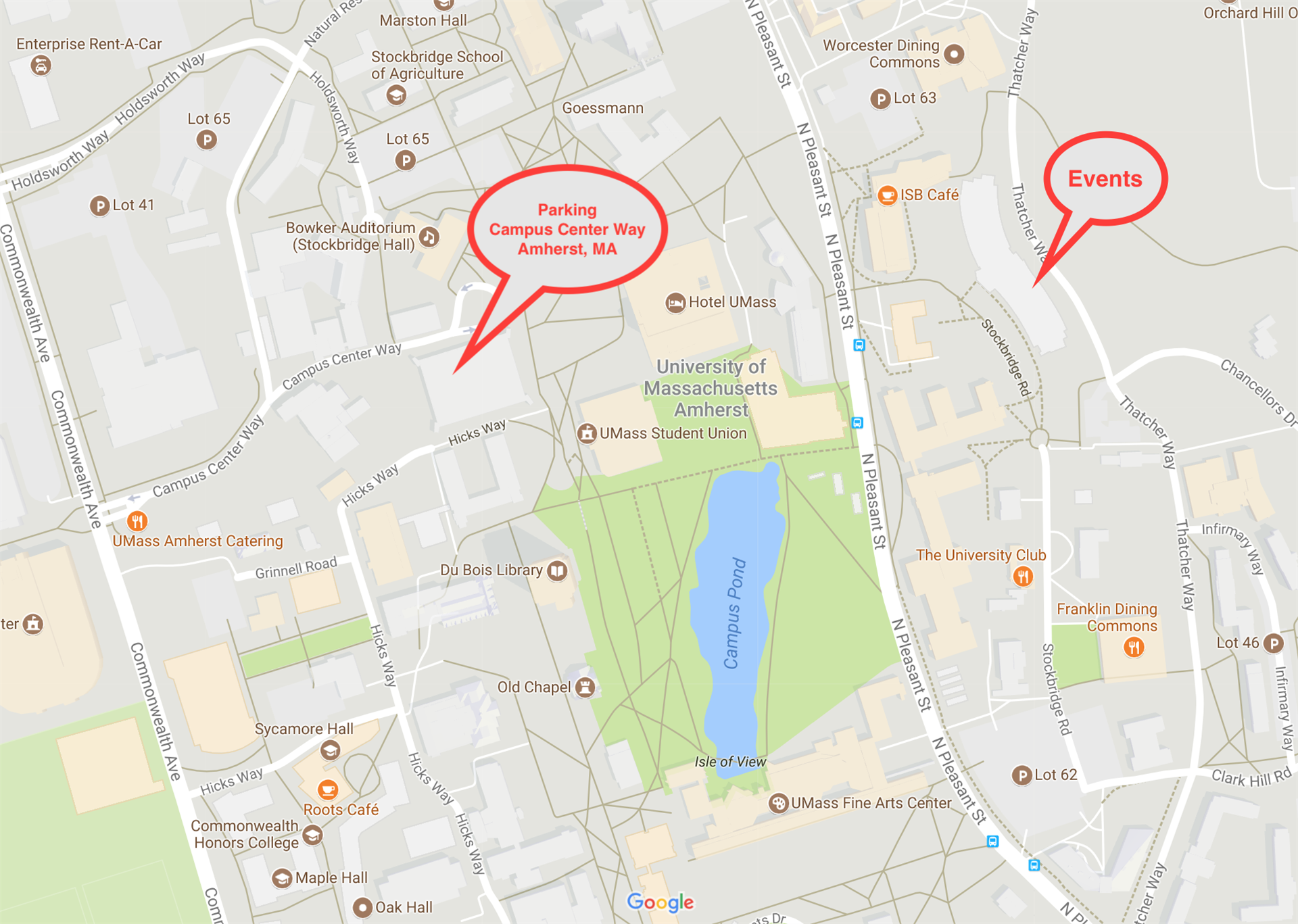
Parking
Campus center garage (pay per hour)
Map "Campus Center Way, Amherst, MA" for directions